We design and discover future biologics, both by establishing novel bispecific formats and by rapidly unveiling new small affinity proteins through phage display and directed evolution.
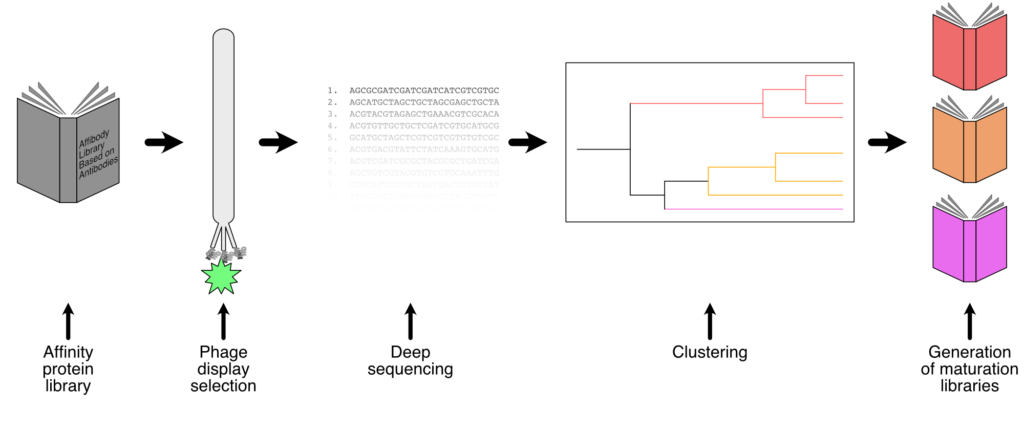
For the discovery of new affinity proteins, we apply our Affibody library (ALBA), which is designed with an antibody inspired diversity at 13 randomized positions. A comprehensive picture of the outcome of the panning rounds is acquired through deep sequencing analysis using in-house developed software. Our program generates maturation libraries based on sequence clusters which are used for a second set of more targeted panning rounds based on the intended application of the binder. Several Affibodies have been isolated from the library towards targets ranging from viral proteins to checkpoint inhibitors, which can later be used in for example bispecific therapeutics.
Multispecific affinity proteins possess a wide variety of potential therapeutic modes of actions. Our work on bispecific formats focuses on the development of bispecific antibodies and a novel bispecific format coined AffimAbs, an antibody-affibody fusion protein, as potential therapeutics within the field of immunotherapy and autoimmune diseases (SLE/RA). Within the scope of the work, we focus on screening for optimal processability and functionality characteristics of bispecifics in a platform-based high-throughput manner. This is done in order to address inherent issues related to the upstream and downstream processing of these complex molecules but also with respect to the targeted disease pathogenesis and the multitude of factors governing it.