The identification and characterization of epitopes, the regions on an antigen that are recognized by antibodies, serves multiple purposes in the development of future cures and treatments. However, being able to in great detail determine conformational epitopes of large and complex proteins can be a laborious undertaking.
In our research group, we have developed a cell display system for epitope mapping using mammalian Chinese hamster ovary cells. The post-translational machinery is highly similar between mammalian cells; therefore, utilizing a mammalian host promises accurately determined epitopes as a high similarity between the protein displayed on the cell surface in our assay is expected to have a high similarity to their natural conformation in human cells.
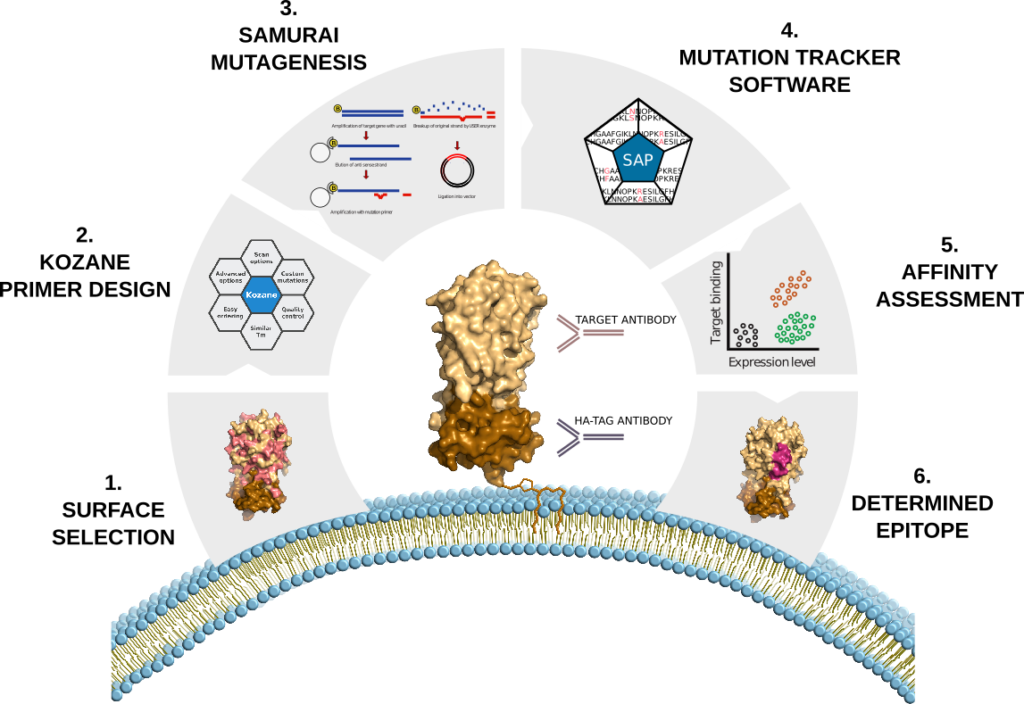
Our workflow for epitope identification is developed with emphasis on speed and reliability. To carry out single amino acid substitutions of surface exposed regions in a high-throughput manner, we apply our in-house methods for mutagenesis (SAMURAI), automated solid-phase cloning, mutagenesis primer design tool Kozane (available at www.kozane.app), and scripts for Sanger sequence evaluation. Generated display vectors with amino acid substitutions are subsequently used to transfect CHO cells and paratope-epitope binding is measured using flow cytometry.
Highlighted research
SAMURAI (Solid-phase Assisted Mutagenesis by Uracil Restriction for Accurate Integration) for antibody affinity maturation and paratope mapping.
Hu FJ, Lundqvist M, Uhlén M, Rockberg J. (2019). Nucleic Acids res. 47(6):e34. 10.1093/nar/gkz050.
This paper presents SAMURAI, our high-thoughput mutagenesis method. It shows how combining a solid-phase support and uracil restriction allows for a reliable and easy-to-use workflow when constructing either multiple single mutants or combinatorial libraries.
Kozane (www.kozane.app)
Kozane is a freely available web application that has been developed to make primer design for site-directed mutagenesis as seamless as possible. It should work for any method that relies on primers with the mismatch in the middle of the oligo, such as SAMURAI, Quikchange®, PFunkel and One-pot Saturation Mutagenesis.
Stratification of responders towards eculizumab using a structural epitope mapping strategy.
Volk AL, Hu FJ, Berglund MM, Nordling E, Strömberg P, Uhlen M, Rockberg J. (2016). Sci Rep. 6:31365. 10.1038/srep31365.
Using a structural epitope mapping strategy based on bacterial surface display, flow cytometric sorting and validation via haemolytic activity testing, we identified six residues essential for binding of eculizumab to C5.